About LesionDiff
Why LesionDiff?
Generating plant disease images with diffusion models is challenging due to lesion variability and limited data. Our approach, illustrated in Fig.1, integrates domain-specific reference data to overcome these challenges, producing images that align closely with human perception and effectively addressing data shortages to enhance disease management.
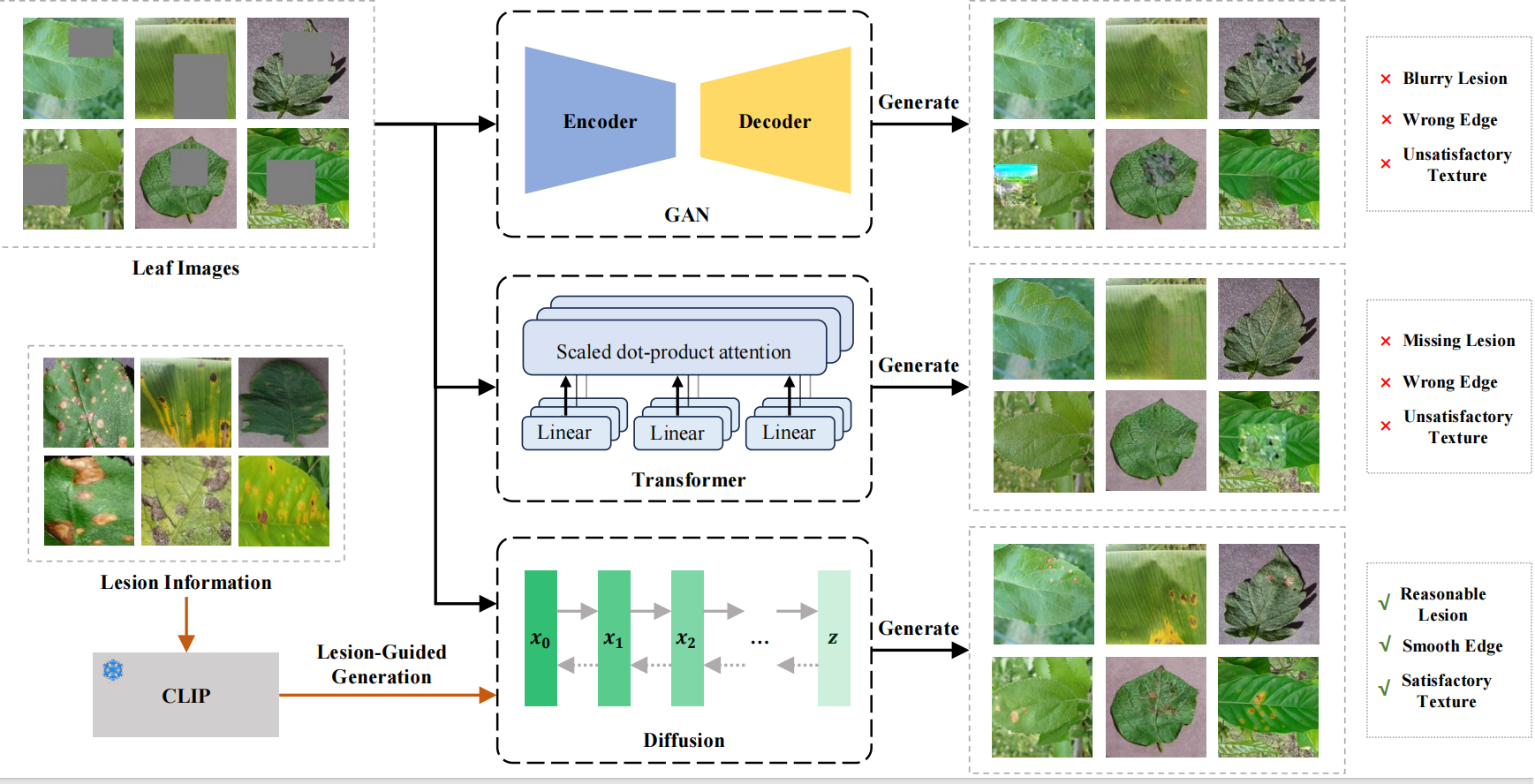
Figure 1: Comparison of generated plant disease images with the original.
Application and Prospects
Diffusion networks like LesionDiff transform plant disease diagnosis by generating diverse, realistic images that enrich training data and improve model accuracy for early disease detection. This advancement supports farmers and agronomists in making proactive management decisions to reduce crop loss and increase yield. Our results demonstrate the promise of diffusion-based models in data-driven agriculture, with future work aimed at extending LesionDiff to more crops and diseases and integrating real-time, scalable detection techniques to tackle evolving plant health challenges.
Conclusion
Accurate plant disease detection is vital for control and yield improvement but requires large datasets that are hard to obtain. To tackle this, we propose LesionDiff, a diffusion-based model that generates realistic synthetic disease images. It includes modules for lesion detection, diversity enhancement, and masked image completion guided by CLIP. Training diagnosis models with both synthetic and real images improves accuracy by over 3%, demonstrating LesionDiff’s effectiveness. Future work will extend its use to more species and real-time synthesis for proactive disease control.
Research Team
SAMLab
