LesionDiff: Measurement System
Introduction
LesionDiff is a diffusion-based framework for generating realistic plant disease images through three main modules: preprocessing, data enhancement, and generation. The preprocessing module uses fine-tuned GroundingDINO to extract lesion areas and create masked images that retain key features. The enhancement module applies a lesion-centered strategy to produce correlated images, boosting dataset diversity. The generation module employs pre-trained CLIP to guide a diffusion model in reconstructing accurate lesion details. Validated by co-training a ResNet18 classifier with both real and generated images, LesionDiff significantly improves disease diagnosis accuracy.
GitHub: https://github.com/GZU-SAMLab/LesionDiff
Environment:
You can create a new Conda environment by running the following command:
conda env create -f environment.yml
In the environment, we still use other networks. If it does not work, please configure the environment of other networks first.
Code and Data
The code can be downloaded from there.
PreTrained Model
The pre-trained PlantDid model is linked below, you can download it.PlantDid: Here are the trained PlantDid weights.
And we use GroundingDINO, the download is: Here.
Get Started
Testing
If you want to batch test, you can use leaf_diseases/inference_bench_gd_auto.py. For example,
python leaf_diseases/inference_bench_gd_auto.py
--config configs/leaf-diseases_with_disease.yaml
--test_path TestDataset/apple
--outdir Result/inferenceTest
--ddim_steps 200 --ddim_eta 1.0 --scale 4 --seed 250
--ckpt models/last.ckpt
--max_size 250
Results
Quantitative Analysis
LesionDiff surpasses other models in producing realistic and varied plant disease images, achieving superior FID and KID scores. By focusing on generating visually convincing lesions instead of exact pixel-level details, it proves highly effective for synthesizing plant disease imagery.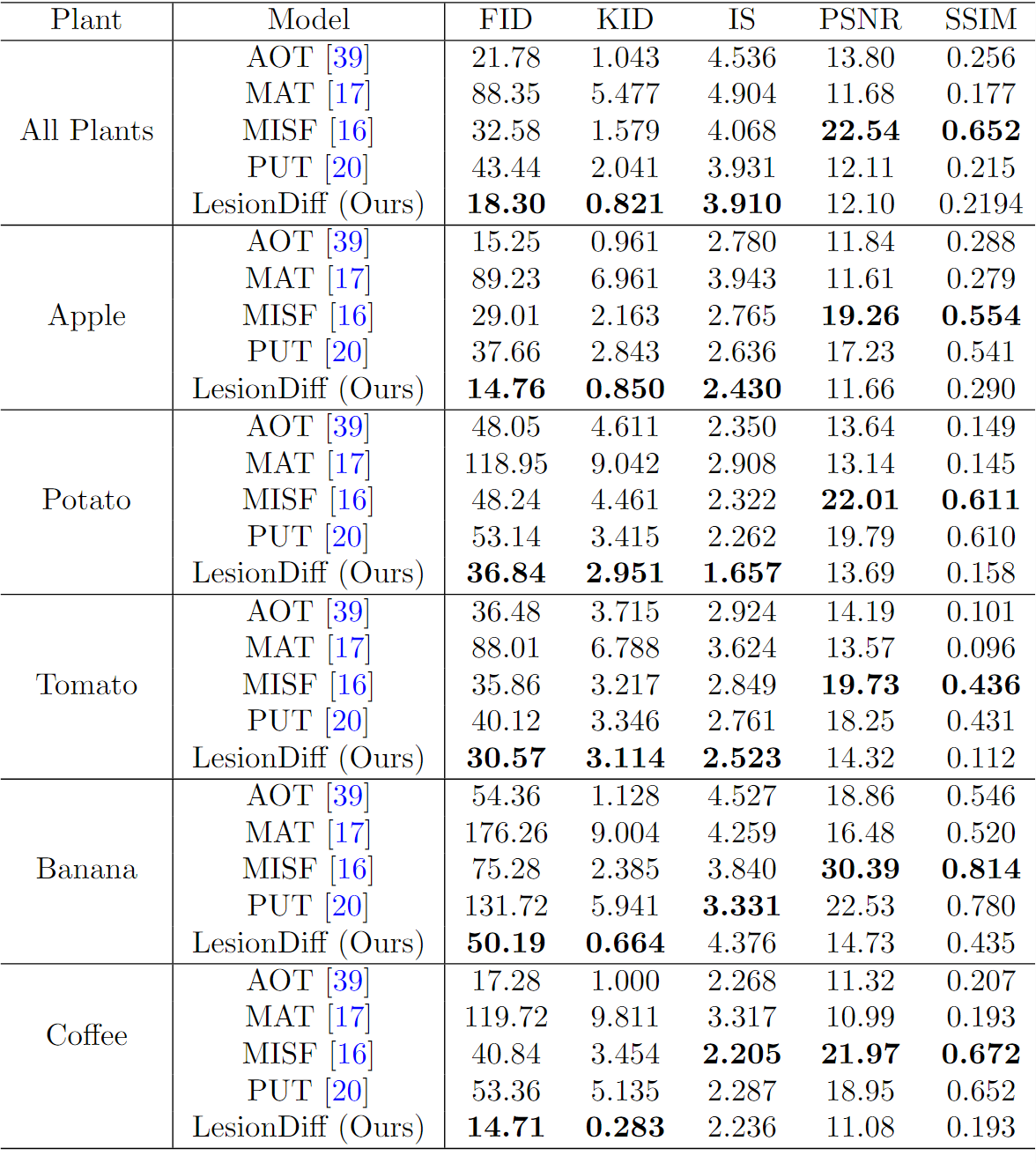
Figure 1:Quantitative evaluation results of LesionDiff and baseline across various plant types.
LesionDiff consistently exceeds baseline models by producing realistic, detailed plant disease lesions across various species and challenging environments, showcasing its robustness and effectiveness for agricultural disease modeling.

Figure 2: Visualization of plant disease images generated by LesionDiff and the baseline model.
Experimental results show that images generated by LesionDiff markedly enhance recognition accuracy, with models trained on this synthetic data surpassing 92% accuracy.
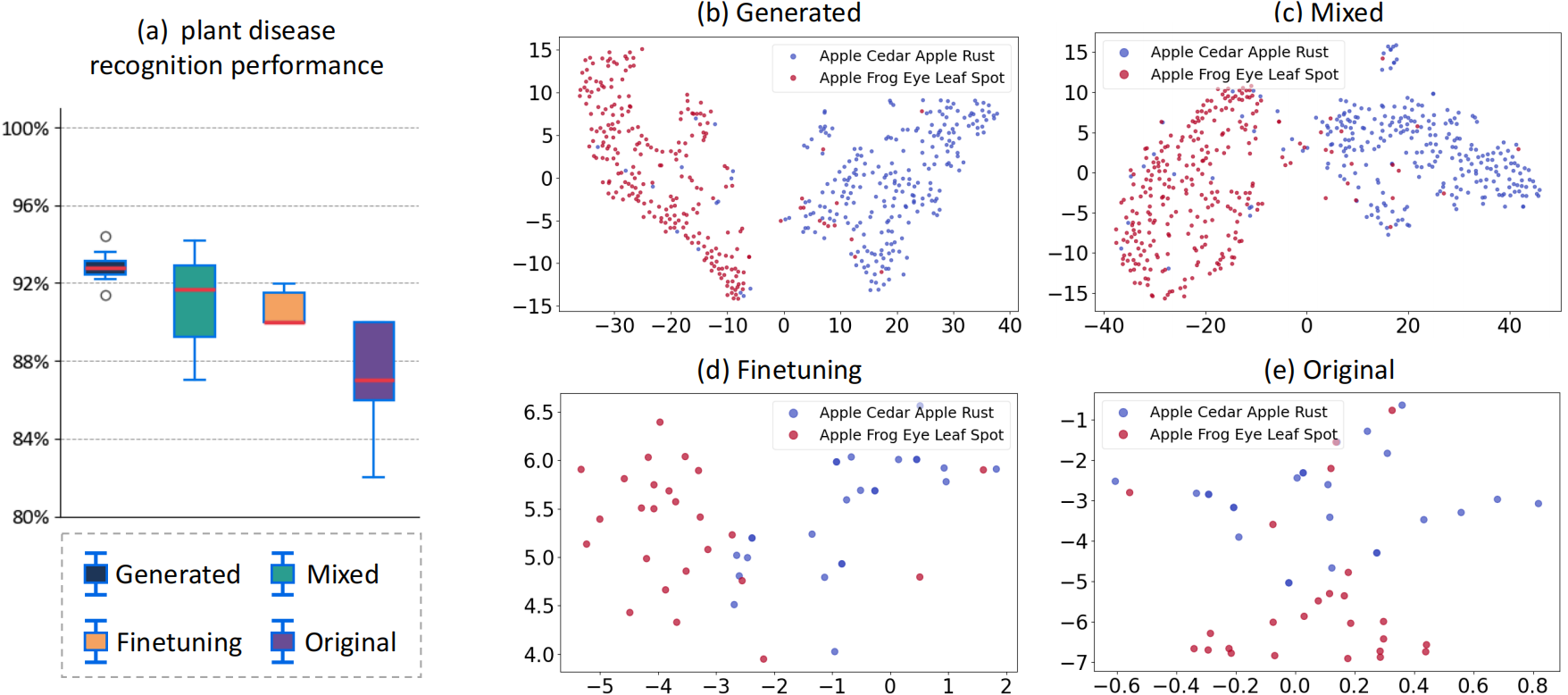
Figure 3: Demonstrate the use of generated images to promote plant disease recognition performance.
